3.2 MeDIP-Seq data analysis for DNA methylation
3.2.1 MeDIP-Seq data analysis for DNA methylation
Single-end reads from MeDIP-sequencing were mapped to the human reference genome (build 19) using the alignment software Eland (version 2) with default parameters. The reads with base quality less than 20 and redundant reads were removed from further analysis. The statistics of mapping MeDIP reads is shown in Supplementary Table 14. The unique reads for each patient was about 15.4 million (37.6%) on average.
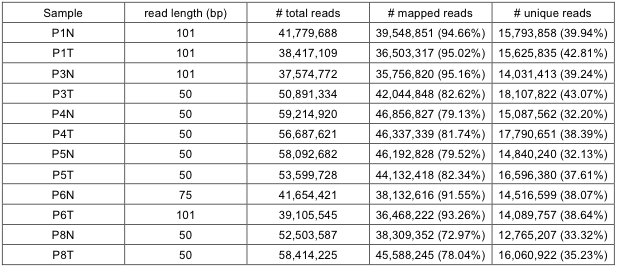
Supp. Table 14. Mapping Summary for MeDIP-Seq data
DNA methylation pattern represents a potentially valuable biomarker in various types of cancer. We identified the differentially methylated regions (DMRs) using edgeR program with the FDR cutoff value of 0.05. All aligned unique reads were extended to be 200 bp long to compensate for the difference in read length and to incorporate potential methylation sites nearby. We did not use the methylation level as additional cutoff for MeDIP-Seq data. In total, we found 558 DMRs, and almost 75% of them were in the promoter or 5’ UTR regions. The list of DMRs with genome annotation (from the UCSC genome browser database for hg19) is provided in the Supplementary File 4.
3.2.2 Integrative analysis of CNV and methylation data with RNA-Seq
Copy number variations and DNA methylation are important factors in regulating gene expression. We investigated the correlation between CNV and DNA methylation with mRNA expression level in a similar fashion to what was done in the microRNAs-target correlation study. For CNV data, we obtained 107 positive correlations with the correlation cutoff of 0.5 (Supplementary Table 12). Highly correlated genes include several genes involved in gene fusion such as STK3, NDUFB9, COX6C, FAM84B, and PTK2. Anti-correlation analysis on DNA methylation in promoter regions yielded 32 relations whose correlation coefficients were smaller than -0.3 (Supplementary Table 13).
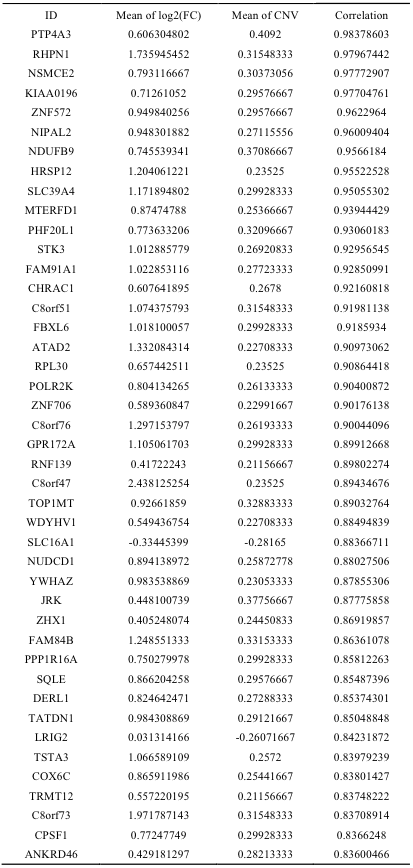
Supp. Table 12. Correlations of gene between CNV and RNA expression
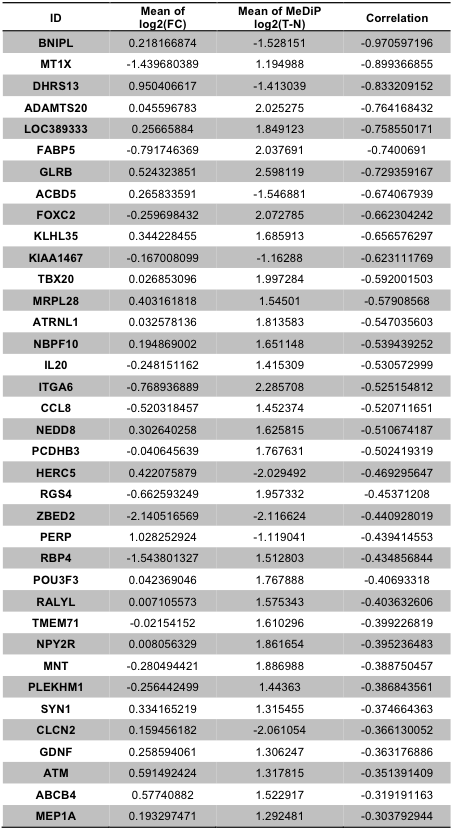
Supp. Table 13. Correlations of gene between DNA methylation and RNA expression